
AVerMedia: Plugins for DarkCrystal HD Capture cards (Windows only) iSight Capture: Webcam video capture using JavaCV and OpenCV.
COLOCALIZATION IMAGEJ MAC
Analysis was performed in at least 3 independent experiments with 10 neurons per condition and 3 ROIs per neuron, using JACoP plugin in ImageJ. Lumenera: Infinity USB 2.0 cameras (Mac only) Dage-MTI: Plugin for XLV, XL16 and XLM cameras (Windows only) Jenoptik: Mac and Windows plugins for ProgRes microscope cameras.
COLOCALIZATION IMAGEJ HOW TO
Description This plugin displays a correlation diagram for two images (8bits, same size). I am analyzing the colocalization of PSD95 and synapsin-1 using ImageJ coloc2 plugin, but I am not sure how to treat the images, such as background subtraction, and the threshold of pictures in. For dynamic structures such as cells, we can observe a change in the amount of colocalization, depending on time or stimulation.
COLOCALIZATION IMAGEJ DOWNLOAD
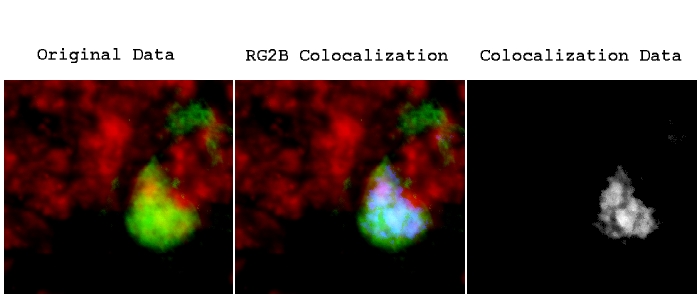
Installation: Download ColocalizationFinder.jar to the plugins folder, or subfolder, restart ImageJ, and there will the Colocalization Finder new command in the Plugins menu. Graph shows quantification of SUMO-1/PSD95 colocalizing pixels. Installation Source: Contained in the JAR file. The colocalized clusters are indicated as yellow puncta. Examples of fluorescence images of dendrites stained for SUMO-1 and PSD95. C) ChemLTP increases SUMO-1 clusters at synapses. Analysis was made in at least 3 different ROIs for each neuron (n = 10), for each condition in 3 independent experiments. Colocalized puncta were quantified using JACoP plugin in ImageJ. Example of dendritic segments stained for SUMO-1 (upper panel) or SUMO-2 (lower panel) protein (blue) and Ubc9 (red). macro recording and execution) Stack Correlation Analyser: Processes a stack image with multiple channels. B) ChemLTP enhances colocalization between SUMO-1 and Ubc9, and SUMO-2/3 and Ubc9 in cultured hippocampal neurons. This is a modified version of the original ImageJ CDA plugin: CDA (macro) Provides support for the CDA plugin within ImageJ macros (i.e. Graphs show quantification of 3 independent experiments, 10 neurons per condition (ANOVA. Restart ImageJ to add the 'RG2BColocalization' command to the Plugins menu. Mean fluorescence intensity was analysed using ImageJ. The presented work disseminates Colocalization Colormap –an ImageJģ plugin that implements the method created by Jaskolski et al.Ģ with the aim to extend its applicability for research.SUMOylation Is Required for Glycine-Induced Increases in AMPA Receptor Surface Expression (ChemLTP) in Hippocampal Neurons Figure 2ĬhemLTP upregulates of SUMO-1 and Ubc9 protein levels.Ī) ChemLTP increases in SUMO-1 and Ubc9 immunoreactivity with no change in the intensity of SUMO-2 staining. Colocalization is then evaluated using these binary images, generally by comparing the area/volume of the intersection of the two images to the area/volume of: a) the union of the binary images, b) the difference of the binary images, c) one of the binary images unaltered, or d) a combination of these three. can be calculated using ImageJ each coefficient has its strengths and. This second part of the plugin proposes to display non-normalized FRET index images in a novel and interactive way to correlate FRET and colocalization between the two fluorophores. Unfortunately, the applicability of the method is limited because there is no computational tool that offers the algorithm among available commercial and public domain software. colocalisation analysis which probably reflects the fact that one approach does. The algorithm was successfully tested on simulated images as well as on biological specimens, becoming an important contribution to colocalization analyses. Here we describe an open source plugin for ImageJ called EzColocalization to visualize and measure colocalization in microscopy images. Most of the algorithms utilized to analyze colocalization correlate global distributions of fluorescence intensities and do not provide any spatial representation of colocalization within the correlated specimens.ġ To overcome this limitation, an advanced colocalization analysis has been proposed by Jaskolski et al.Ģ This inventive algorithm generates a map of correlations between pairs of corresponding pixels in the two investigated images which allows for both quantification and visualization of colocalization. This plugin contains most of existing colocalization methods in 2D and 3D fluorescence microscopy images: Pearson and Manders coefficients, Image Cross Correlation Spectroscopy (ICCS) and Object-based methods. ExcerptSeparate fluorescent labelling with different fluorophores is a typically used protocol to determine the colocalization of the two distinct biological elements in the overlapping microscopic images.


 0 kommentar(er)
0 kommentar(er)
